Scientists from the Novo Nordisk Foundation Center for Biosustainability (DTU) have found four new anti-CRISPR proteins that are distributed across different environments. The new study published in Cell Host & Microbe suggests that some anti-CRISPR proteins are more widespread in nature than previously anticipated. These anti-CRISPRs can potentially be used to regulate the activity of CRISPR-Cas9 systems better in the future.
CRISPR systems are bacterial immune systems that enable the bacterium to fight off infecting viruses (phages) in a targeted manner.
Due to their programmable nature CRISPR systems, and in particular Cas9, are currently being widely deployed in the life science industry with the potential to deliver breakthrough gene therapies, new antibiotics and malaria therapies.
Interestingly, phages have evolved anti-CRISPR proteins to overcome bacterial CRISPR systems in the evolutionary arms race between viruses and bacteria. These proteins quickly inhibit the host bacterium’s defence system leaving the bacterium vulnerable to infection.
In spite of their significant biological importance, only a few anti-CRISPR proteins have been discovered so far in a very specific subset of bacteria. Current anti-CRISPR proteins are not abundant in nature and have been identified by studying the DNA of the phages that were able to infect bacteria harbouring CRISPR-Cas9. Using this method, one relies on being able to culture bacteria and on phages that are able to infect and avoid the surveillance of the endogenous CRISPR Cas9-system.
Find your dream job in the space industry. Check our Space Job Board »
“We used a different approach that focused on anti-CRISPR functional activity rather than DNA sequence similarity. This approach enabled us to find anti-CRISPRs in bacteria that can’t necessarily be cultured or infected with phages. And the results are really exciting,” says Ruben Vazquez Uribe, Postdoc at the Novo Nordisk Foundation Center for Biosustainability (DTU).
Faecal samples contained anti-CRISPRs
The researchers identified the anti-CRISPR genes by using the total DNA from four human faecal samples, two soil samples, one cow faecal sample and one pig faecal sample. The DNA was chopped into smaller pieces and randomly expressed on a plasmid within a bacterial cell. This cell contained a genetic circuit for selection of anti-CRISPR activity. In short, this meant that cells containing a plasmid with a potential anti-CRISPR gene would become resistant to a certain antibiotic. On the contrary, cells in which the plasmid did not confer anti-CRISPR-activity would die.
With this system, the researchers could easily detect and select DNA with anti-CRISPR activity and trace it back to its origin. Using this metagenomic library approach, the scientists were able to identify eleven DNA fragments that circumvented Cas9 activity.
Further characterisation could then confirm the activity of four new anti-CRISPRs. Phylogenetic analysis revealed that the genes identified in the faecal samples are present in bacteria found in multiple environments, for instance in bacteria living in insects’ gut, seawater and food.
This shows that the newly discovered genes are spread in many bacterial branches in the tree of life, and in some cases with evidence that some of these genes have been horizontally transferred numerous times during evolution.
“The fact that the anti-CRISPRs we discovered are so abundant in nature suggest that they are very useful and have a big significance from a biological perspective,” says Morten Sommer, Scientific Director and Professor at the Novo Nordisk Foundation Center for Biosustainability (DTU).
These findings suggest that anti-CRISPRs could likely play a much bigger role in the interplay between phage and host than has previously been suggested.
Useful switch for genome editing
Earlier studies in this field have demonstrated that anti-CRISPR proteins can be used to reduce errors, such as cutting DNA at off-target sites, when doing genome editing in the laboratory.
“Today, most researchers using CRISPR-Cas9 have difficulties controlling the system and off-target activity. Therefore, anti-CRISPR systems are very important, because you want to be able to turn your system on and off to test the activity. Therefore, these new proteins could become very useful,” Morten Sommer says.
The researchers actually discovered that the four new anti-CRISPR proteins seem to have different traits and properties. Going forward, this will be very exciting to investigate further.
Provided by:
Technical University of Denmark
More information:
Ruben V. Uribe et al. Discovery and Characterization of Cas9 Inhibitors Disseminated across Seven Bacterial Phyla. Cell Host & Microbe (2019). DOI: 10.1016/j.chom.2019.01.003
Image:
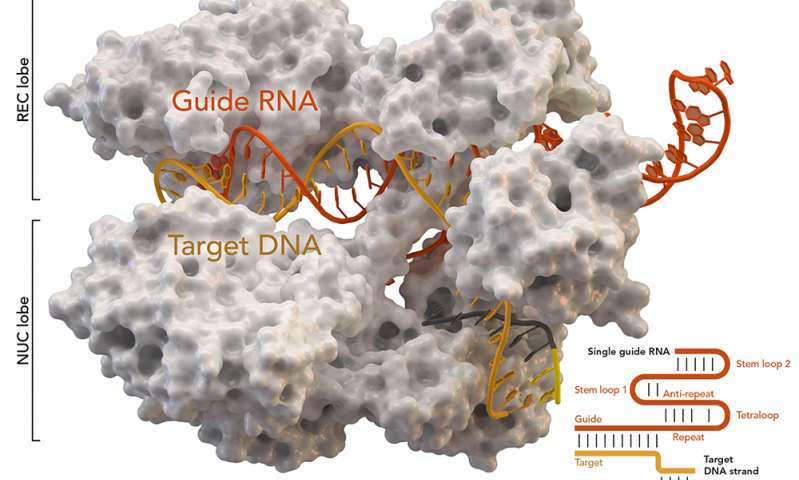
CRISPR-associated protein Cas9 (white) from Staphylococcus aureus based on Protein Database ID 5AXW.
Credit: Thomas Splettstoesser (Wikipedia, CC BY-SA 4.0)