Porphyromonas gingivalis is a major bacterial pathogen that leads to periodontitis, also known as gum disease. In Japan, 80% of adults aged 35 and over suffer from this disease. What’s more, P. gingivalis has also been linked to rheumatoid arthritis, cardiovascular disease, pancreatic cancer and even Alzheimer’s disease.
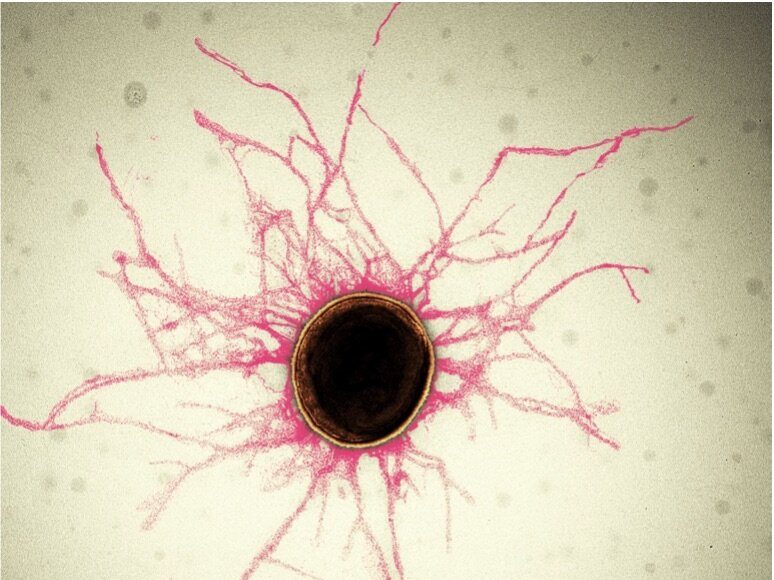
Periodontitis is an oral inflammatory disease in response to biofilms—a bacterial plaque that accumulates on surfaces like our teeth. Biofilms are primarily created by bacterial cells attaching themselves to the host and to each other via sticky, hair-like filaments called pili. In serious cases, periodontitis can result in gum erosion and tooth loss.
A team of researchers from the Molecular Cryo-Electron Microscopy Unit at the Okinawa Institute of Science and Technology Graduate University (OIST), alongside the groups of Professor Koji Nakayama at Nagasaki University and Professor Katsumi Imada at Osaka University, have revealed the structure of these adhesive pili and shed light on how they assemble. Their research, published in Nature Microbiology, has provided new insights into bacteriology and is a crucial step towards combating the diseases with which this bacterium is associated.
“Pili are vital for both the survival of the bacteria, and the creation of the biofilms,” said Dr. Satoshi Shibata, first author and staff scientist in OIST’s Unit, which is led by Associate Professor Matthias Wolf. “By taking a close look at these pili, our research has provided insights into how we can prevent biofilms from forming.”
P. gingivalis is a member of the class Bacteroidia. Previous research, led by Professor Nakayama and colleagues at Nagasaki University, found that most bacteria within this class have unique Type V pili. However, until now, the structure and assembly process of these pili were unknown. “Besides periodontal pathogens, Type V pili are seen in major colon bacteria such as Bacteroides and Prevotella species and their Type V pili may contribute to formation of colon microbiota,” said Professor Nakayama.
Find your dream job in the space industry. Check our Space Job Board »
Pili themselves are made up of smaller protein units called pilins. In the case of P. gingivalis pili, most of these are FimA pilins. Although pilins are only linked by weak interactions, they can assemble into very stable pili.
The first step to determining how this assembly occurred was to take a close look at the structure of individual pilins. “Detailed structural information of FimA is very important because pathogenicity of P. gingivalis strains is closely related with the FimA sub-types,” said Professor Imada.
Professor Imada and students from Osaka University crystalized FimA pilins, revealing their unassembled state at atomic resolution.
“Based on findings from earlier experiments by the Nakayama group, we theorized that these pilins assembled themselves via a mechanism of protease-mediated strand-exchange,” said Dr. Shibata. “So, our next experiment took a close look at fully assembled pili using cryo-electron microscopy.”
Associate Prof. Mikio Shoji from the Nakayama group, and Dr. Shibata prepared a genetically engineered version of the FimA pilins, which successfully assembled into pili, after a protease—a protein that cuts other proteins—was added. Dr. Shibata then collected thousands of images on the high-end cryo-electron microscope at OIST and processed the data on the University’s “Sango” supercomputer, resulting in a complete three-dimensional atomic model of the assembled pilus structure.
“When we added the protease, the pilins started to assemble into elongated pili like train cars connecting to form a train. This happened because the protease cut a retaining loop and released a protein strand, known as the donor strand, which triggered the assembly to begin,” said Professor Wolf. Once released, the donor strand flipped out of the pilin and inserted itself into a neighboring pilins groove, thus connecting the two pilins.
Finally, in a combined team effort, the three groups took a closer look at the amino acid composition at the end of the donor strand and found that it played a critical role in the assembly mechanism. Using biochemistry, crystallography and cryo-EM, they mutated the protein, which prevented the pili from forming and thus proved how these key amino acids contribute to pilin polymerization.
Ultimately, this research is a step towards new anti-bacterial drugs, not just for the diseases caused by P. gingivalis, but for those caused by any bacteria containing Type V pili. “We’re now trying to create an inhibitor that prevents pili from assembling,” said Dr. Shibata. “This structure serves as a target to create new drugs, which are desperately needed to counter increasing antibiotic resistance. Finding novel antimicrobial compounds is a critical advantage in fighting these pathogens.”
Provided by: Okinawa Institute of Science and Technology
More information: Satoshi Shibata et al. Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange. Nature Microbiology (2020). DOI: 10.1038/s41564-020-0705-1
Image: Most bacterial cells are covered in tiny, hair-like structures called pili. Their diameter is smaller than 1/10,000th of a human hair.
Credit: S. Shibata, OIST