DNA is tightly packed into the nucleus of a cell. Nevertheless, the cellular machinery needs to constantly access the genomic information. An LMU team now reveals the inner workings of a molecular motor made of proteins which packs and unpacks DNA.
The genomic DNA of higher organisms is compacted in a highly condensed form known as chromatin. The DNA is tightly wound around a myriad of tiny histone spools called nucleosomes. A single human cell, for instance, accommodates in this manner about two meters of DNA. However, genes must be constantly transcribed into messenger RNAs to direct protein synthesis. Moreover, the entire DNA must be replicated before cell division and DNA damage needs to be repaired. Thus, there must be way to actively grant access to the genome.
This is when chromatin remodelers come into play. Chromatin remodelers have an essential role as they are molecular machines: they unpick and unpack segments of the DNA by sliding nucleosome spools back and forth, replacing individual histones, freeing up the DNA for transcription, and finally compacting it again, when the job is done. Since all of this happens in a highly dynamic fashion, chromatin remodelers enable cells to react rapidly to alterations in their environment – and this holds for brewer’s yeast as well as for human cells. In mediating gene accessibility, chromatin remodelers are vital for development and cell differentiation; cell types are defined by the sets of genes they express, remodelers help to determine cell identity.
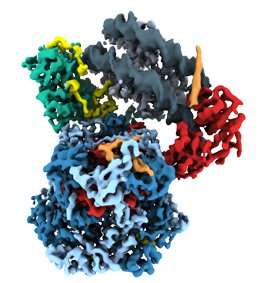
So far, however, very little is known about what remodeling proteins look like and how they go about doing what they do. In molecular terms, functional remodelers are often very large complexes comprising many different protein components, whose coordinated action makes them akin to molecular machines. These features also make it very difficult to determine their detailed structure. But a team led by Professor Karl-Peter Hopfner, who holds a Chair in Structural Molecular Biology at LMU’s Gene Center, has now used cryo-electron microscopy to reconstruct the three-dimensional structure of the nucleosome-sliding remodeler INO80 (which itself consists of 15 subunits) bound to a single nucleosome. “Even with innovative approaches, the best available technology and intensive teamwork, we were always working at the cutting edge,” says Dr. Sebastian Eustermann, who worked out the molecular structure of the complex on the basis of electron micrographs of thousands of individual complexes.
By analyzing images of randomly oriented views of the complex formed between INO80 and a nucleosome in the electron micrographs, Hopfner and his team have pieced together its structure at a resolution which has seldom been achieved for a chromatin complex of comparable size. This allowed the researchers to unravel the intricate interaction of the remodeler with its substrate DNA spooled around histones and dissect how the whole machinery works.
Find your dream job in the space industry. Check our Space Job Board »
From a biochemical point of view, remodelers are responsible for heavy-duty reorganizational tasks. To perform these tasks, they must execute “large-scale conformational changes, which are carried out with astounding precision,” says Eustermann. In order to alter the relative positions of nucleosomes, the INO80 complex must first weaken the contacts between the nucleosomal histones and the DNA. A molecular motor which is part of the INO80 complex segmentally detaches the double-stranded DNA from the nucleosome. In doing so, it progressively breaks the contacts that normally keep the DNA tightly wound around the histone particle.
The motor subunit feeds DNA it into the nucleosome. This results in the transient formation of a double-stranded DNA loop that is likely an important intermediate in complex remodeling reactions on the nucleosome. On one hand, the loop exposes some histone proteins that could be replaced by other histones to form a different type of nucleosome. On the other hand, the loop is eventually passed over another subunit and the machine then acts as a ratchet, allowing the nucleosome to “move” on the DNA. Throughout this unpacking process, other subunits in the complex serve to support and stabilize the partially ‘denuded’ nucleosome itself.
The structure of the complex revealed in the new study sheds new light on the function and mode of action of chromatin remodelers in general. These molecular machines play an essential part in the workings of the cell by maintaining the flexibility of the chromatin, thus enabling the genetic apparatus to respond dynamically to changing metabolic demands. “Our results provide the first well-founded picture of how they do that,” says Hopfner. “Moreover, it has recently become clear that remodelers play a central role in tumorigenesis, because they often misregulated in tumor tissue. So structural and mechanistic insights into their functions will be vital for the future development of new therapies for cancer,” he adds.
Provided by:
Ludwig Maximilian University of Munich
More information:
Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature. (2018) DOI: 10.1038/s41586-018-0029-y
Image:
he architecture of INO80’s nucleosome remodeling core
Credit: Hopfner Group