For decades, greater than 60% of the human genome was believed to be “junk DNA” that served little or no purpose in the course of human development. Recent research by Colorado State University is challenging this notion to show that junk DNA might be important after all.
A new study, published on June 5 in Aging Cell, found that a portion of noncoding genetic material, called repetitive element transcripts, might be an important biomarker of the aging process.
Tom LaRocca, an assistant professor in the Department of Health and Exercise Science and faculty member in the Columbine Heath Systems Center for Healthy Aging at CSU, led the study to investigate a growing body of evidence that repetitive elements—transposons and other sequences that occur in multiple copies in the human genome—may become active over time as we age.
LaRocca, graduate student Alyssa Cavalier, and postdoctoral researcher Devin Wahl centered specifically on RNA transcripts, molecules that are transcribed from the DNA of repetitive elements, to test whether they increase in number with age.
“The biomarker angle is important here,” LaRocca said. “Ten to 20 years from now, we might be able to take samples or certain measurements from people in the doctor’s office and get some insight into what’s going on with them biologically, so that we can know how to best treat them and maximize their healthspan. If these repetitive element RNAs are a biomarker of aging, then maybe someday you can get a measurement like this done to see how your repetitive elements are being expressed. Are there too many of them? Is that a problem?”
Find your dream job in the space industry. Check our Space Job Board »
Mapping RNA
To carry out the study, the researchers began by analyzing an existing RNA sequencing dataset gathered from skin cells in healthy human subjects aged 1-94 years old. Just as the Human Genome Project of the 1990s sought to sequence and map the approximately 20,500 genes in human DNA, RNA sequencing can provide a map of the entire transcriptome in the cells under study. From that analysis, which was all computational, the researchers found that transcripts from most major types of repetitive elements were increased in older subjects.
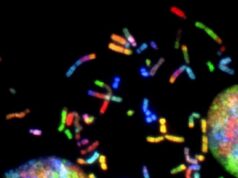
In a second wave of study, the researchers verified their initial findings by performing their own lab analyses on skin cells from a biobank. Using fluorescent microscopy, the researchers tagged the transcript of a specific transposon, Charlie5, to see how it fluctuates with the age of cells: the brighter the tag appears under the microscope, the more Charlie5 transcript is detectable.
As hypothesized, skin cells from older adults revealed a marked accumulation of Charlie5 transcript compared to cells from younger individuals, showing that repetitive element RNAs appear to accumulate with age.
While an important observation, the grander outcome of this study is that repetitive RNA transcripts might be linked with biological age, or the health of a person’s cells, as opposed to chronological age in years.
“If you find something that changes progressively with aging, that finding alone is not necessarily interesting, because lots of things increase or decrease with age. What you really want to find is something that reflects biological aging,” LaRocca said. “For example, let’s say you’re a smoker and you’re under a lot of chronic stress. Then, perhaps even if you’re only 45, your biological age—the health of your cells—could actually be 60 or 65. We think that repetitive element transcripts could be a marker of this.”
Link to aging
To study biological age, Cavalier performed an analysis that compared sun-exposed skin cells to skin cells that had not been exposed to sunlight—the theory being that the more damaging UV rays a skin cell is exposed to, the older the cell will be biologically. Consistent with her hypothesis, Cavalier noted higher levels of repetitive element RNAs in the sun-exposed cells.
A link between repetitive element transcripts and biological age was further confirmed by studying skin cells from patients with Hutchinson-Gilford progeria syndrome (HGPS), a premature aging syndrome, and by studying an RNA-sequencing dataset from the roundworm Caenorhabditis elegans.
Why might repetitive element transcripts increase with age? The researchers suspect that chromatin—the complex of DNA and protein in cells that typically represses repetitive elements from being expressed—might become disrupted, allowing for the transcription of repetitive elements.
All in all, for a portion of the genome that scientists used to ignore, evidence is growing that noncoding RNAs and repetitive elements play vital roles in regulating the rest of the human genome, and in this case, as potentially targetable biomarkers of aging.
“This is a really big chunk of the genome that, for the longest time, no one really knew what it did, so they just kind of assumed it was junk. But we’re finding more and more that these noncoding regions might not only be doing something, but they might have actual health implications,” Cavalier said.
Future studies in LaRocca’s Healthspan Biology Lab will compare chromatin structure in people who exercise routinely with those who don’t to understand how exercise impacts repetitive element levels. Other studies will investigate the possibility of using a drug to inhibit repetitive element RNAs from being transcribed.
Provided by: Colorado State University
More information: Thomas J. LaRocca et al. Repetitive elements as a transcriptomic marker of aging: Evidence in multiple datasets and models. Aging Cell (2020). DOI: 10.1111/acel.13167
Image: Fluorescence microscopy images of the Charlie5 transcript in young versus old human skin cells.
Credit: Aging Cell