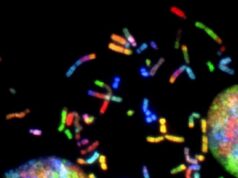
A “navigational map view” of a section of a mouse brain. Like the map view of an Earth imaging program, this image of a brain section takes cues from actual imaging performed with highest-energy X-rays at a synchrotron and turns them into a graphic depiction. The imaging concentrates on a mesa-scale of the brain analogous to the map view of Google Earth. The scale could be useful in studying how the brain computes.
Credit: Georgia Tech / Eva Dyer
If brain imaging could be compared to Google Earth, neuroscientists would already have a pretty good “satellite view” of the brain, and a great “street view” of neuron details. But navigating how the brain computes is arguably where the action is, and neuroscience’s “navigational map view” has been a bit meager.
Now, a research team led by Eva Dyer, a computational neuroscientist and electrical engineer, has imaged brains at that map-like or “meso” scale using the most powerful X-ray beams in the country. The imaging scale gives an overview of the intercellular landscape of the brain at a level relevant to small neural networks, which are at the core of the brain’s ability to compute.
Find your dream job in the space industry. Check our Space Job Board »
Dyer, who recently joined the Georgia Institute of Technology and Emory University, also studies how the brain computes via its signaling networks, and this imaging technique could someday open new windows onto how they work.
Highest-energy X-rays
A powerful X-ray tomography scanner allowed the researchers to image particularly thick sections of the brains of mice, which afforded them views into intact neural areas much larger than are customary in microscope imaging. The scanner operated on the same basic principle as a hospital CT scanner, but this scan used high-energy X-ray photons generated in a synchrotron, a facility the size of dozens of football fields.
“Argonne National Laboratory (ANL) generates the highest-energy X-ray beams in the country at its synchrotron,” said Dyer, who co-led the study with ANL’s Bobby Kasthuri at the Advanced Photon Source synchrotron. “They’ve studied all kinds of materials with really powerful X-rays. Then they got interested in studying the brain.”
The technique also revealed capillary grids interlacing brain tissues. They dominated the images, with cell bodies of brain cells evenly speckling capillaries like pebbles in a steel wool sponge.
“Our brain cells are embedded in this sea of vasculature,” said Dyer, an assistant professor in the Wallace H. Coulter Department of Biomedical Engineering at Georgia Tech and Emory.
The study on the new images appeared in the journal eNeuro on Tuesday, October 17, 2017. The team included researchers from Johns Hopkins University, the University of Chicago, Northwestern University, the Argonne National Laboratory, and the University of Pennsylvania. The work was funded by the U.S. Department of Energy, the National Institutes of Health, the Intelligence Advanced Research Projects Activity, and the Defense Advanced Research Projects Agency.
Neural forest for the trees
Electron microscopy already captures neuronal details in impressive clarity. Functional magnetic resonance imaging (fMRI) makes great visuals of brain structures and broad neural signaling.
So, why do researchers even need mesoscale imaging?
“FMRIs image at a high level, and with many microscopes, you’re zoomed in too far to recognize the forest for the trees,” Dyer said. “Though you can see a lot with them, you also can miss a lot.”
“If you look at brain signaling on the level of individual neurons, it looks very mysterious, but if you take a step back and observe the activity of a population of hundreds of neurons instead, you might see simpler, clearer patterns that intuitively make more sense.”
In an earlier study, Dyer discovered that hand motion directions corresponded with reliable neural signaling patterns in the brain’s motor neocortex. The signals did not occur across single neurons or a few dozen but instead across groups of hundreds of neurons. Mesoscale imaging reveals a spatial view on that same order of hundreds of neurons.
Megamap dreams
The researchers have also been able to couple their new meso-level imaging technique with extremely detailed electron microscopy. And that has the potential to take them closer to a kind of Google Earth for the brain by combining mesoscale or map-like views with zoomed-in or street-like views.
“We have begun doing X-ray tomography on large brain tissues, then we’ve gone deeper into specific tiny regions of interest in the same tissue with an electron microscope to see the full connectome there,” Dyer said. The connectome refers to the total scheme of the hundreds of individual connections between neurons.
The researchers hope to someday be able to switch from a mesoscale view to close-up view, a bit like Google Earth.
Zeroing in then zooming in
“I think what we’re going to need in neuroscience is this ability to traverse across different scales,” Dyer said. She envisions a future multi-scale imaging technology that is useful in understanding neurological diseases.
“We want to be able to tell somebody researching a disease what the underlying anatomy of their lab sample is in an automated way,” she said. “You could navigate using this mesoscale view to get the context of where the damage is.”
Then the user could zoom in on a blocked artery or destroyed tissue analogous to the way satellite imagery can zoom in on traffic jams to see what’s causing them.
From X-ray to graphic image
Like a navigational map, the final images in the study were colorful, clear, mesoscale graphic depictions. They were based on the X-ray tomography, but a lot was involved in getting from the X-ray to the image.
First, the thick section of brain rotated in the high-energy X-ray beam, which was transformed into an image analogous to the output of a CT scanner. Then structures and characteristics were identified by humans and algorithms before they were computed into three-dimensional, color-coded vasculature and cell bodies.
The details of individual cells were very basic. In neurons, often the nuclei were visible in the X-ray tomography image, and axons wrapped in myelin (white matter) sometimes appeared as well.
Pragmatic computation
The new mesoscale imaging of brain samples also has pragmatic advantages.
It may be possible to examine minuscule brain regions piece by piece with electron microscopes then compute them together into a complete image of the brain, but it’s hardly practical. “Producing a three-dimensional map of just a cubic millimeter of the brain with an electron microscope requires processing petabytes of data,” Dyer said.
By contrast, the researchers need 100 gigabytes of data to compute a one-cubic-millimeter image of brain tissue using mesoscale X-ray tomography scans of thicker brain sections. But the researchers’ goal is to not have to slice the tissue at all.
“Eventually, we want to be able to image whole brains, as is, with this method to see the entirety of their neural networks and other structures.”
This study was co-authored by William Gray Roncal and Joshua T. Vogelstein of Johns Hopkins University, Judy Prasad of the University of Chicago, Hugo L. Fernandes of Northwestern University; Doga Gürsoy, Vincent De Andrade, Kamel Fezzaa and Xianghui Xiao of Argonne National Laboratory; Chris Jacobsen of Argonne and Northwestern, and Konrad Körding of the University of Pennsylvania. Research was funded by the U.S. Department of Energy Office of Science User Facilities operated by Argonne National Laboratory (contract DE-AC02-06CG11357), the National Institute of Mental Health at the National Institutes of Health (grant U01MH109100), the Intelligence Advanced Research Projects Activity MICrONS project, the Defense Advanced Research Projects Agency SIMPLEX program (contract N66001-15-C-4041) and DARPA GRAPHS program (contract N66001-14-1-4028).
Story Source: Materials provided by Georgia Institute of Technology. Original written by Ben Brumfield. Note: Content may be edited for style and length.
Journal Reference:
Eva L. Dyer, William Gray Roncal, Judy A. Prasad, Hugo L. Fernandes, Doga Gürsoy, Vincent De Andrade, Kamel Fezzaa, Xianghui Xiao, Joshua T. Vogelstein, Chris Jacobsen, Konrad P. Körding, Narayanan Kasthuri. Quantifying Mesoscale Neuroanatomy Using X-Ray Microtomography. eneuro, 2017; 4 (5): ENEURO.0195-17.2017 DOI: 110.1523/ENEURO.0195-17.2017