In a joint collaboration, Danish and German researchers have characterized a cellular activity that protects our cells from potentially toxic by-products of gene expression. This activity is central for the ability of multicellular organisms to uphold a robust evolutionary ‘reservoir’ of gene products.
Manufacturing processes need quality control systems in order to ensure proper assembly of functional products. Moreover, space-consuming, and perhaps even toxic, by-products of such processes need to be properly discarded or recycled by efficient waste handling systems.
By analogy, transcription of our genome is an imperfect process that produces large quantities of non-functional and potentially harmful transcripts both from within and outside of conventional genes. The RNA polymerase II enzyme transcribes the majority of our genes, and it also generates pervasive transcripts from multiple non-genic regions. Over the past decade, it has become increasingly clear that the enzyme is relatively promiscuous when it comes to where it starts, and there are transcription initiation sites scattered everywhere in the genome. However, not all these sites are associated with a ‘proper’ gene, and this requires the presence of specific gene-defining elements.
Discovery of new activity of the protein complex ‘Integrator’
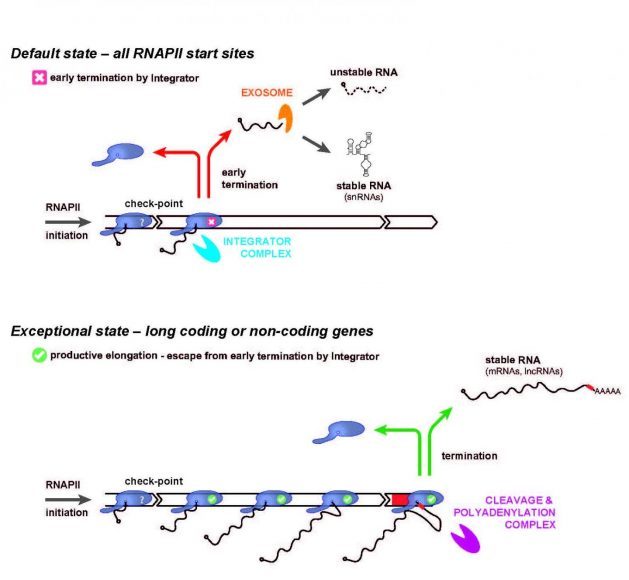
In a new article published in the international journal Molecular Cell, a Danish-German research team has now demonstrated that the multi-protein complex ‘Integrator’—which was previously described as the transcription termination factor for a specific class of genes encoding small nuclear (sn)RNAs—is in fact a default ‘early’ termination factor for most, if not all RNA polymerase II initiation events (Figure, top panel).
Find your dream job in the space industry. Check our Space Job Board »
The resulting short transcripts are in most cases rapidly degraded by the ribonucleolytic RNA exosome. This mechanism is also at play to varying degrees inside conventional genes, but these have evolved elements, counteracting such early termination, to facilitate productive transcription elongation, which eventually produces functional transcripts such as protein-coding ‘messenger’ (m)RNA (Figure, bottom panel).
Through this newly discovered mechanism of action, Integrator ensures that production of wasteful transcripts remains limited, while at the same time allowing maintenance of thousands of transcription start sites under neutral selection in the human genome. These provide a reservoir of transcription units that may turn into functional genes over time, exemplified by the evolution of the currently known functional RNAs of our genome. As a curious example, snRNA genes appear to be a special case, taking advantage of the general early termination activity of Integrator and fend off the ensuing RNA exosome activity to allow production of stable functional RNAs (Figure, top panel).
Inactivating mutations in Integrator subunits lead to severe neurodevelopmental disorders, and elevated expression of some integrator subunits is associated with increased epithelial-to-mesenchymal transition—a key step in tumor metastasis. This duality highlights the need for a tight control of the Integrator activity. Moreover, the realization that Integrator is a general attenuator of nonproductive transcription may inform the molecular characterization and potential treatment of such ailments.
Provided by: Aarhus University
More information: Søren Lykke-Andersen et al. Integrator is a genome-wide attenuator of non-productive transcription. Molecular Cell (2020). DOI: 10.1016/j.molcel.2020.12.014
Image: RNA polymerase II (RNAPII) initiates transcription at a multitude of positions in the human genome. However, only a fraction of these sites is associated with genes that give rise to functional RNA. The ‘Integrator’ complex by default terminates RNAPII shortly after its initiation (top), unless specific gene-defining elements are present allowing for transition to productive elongation (bottom). In the latter case, transcription is typically terminated by the ‘Cleavage & Polyadenylation’ Complex that recognizes specific ‘terminator’ sequence elements in the genome (bottom, red box). In the former case the produced short RNA is by default rapidly degraded by the RNA exosome, whereas snRNAs constitute a special case that avoids degradation.
Credit: Søren Lykkke-Andersen